Computational biology has emerged as an indispensable discipline at the intersection of biological research & computer science, primarily focusing on biomolecular structure prediction. The ability to accurately predict these structures has profound implications for understanding cellular functions and developing new medical therapies. Despite the complexity, this field is pivotal for gaining insights into the intricate world of proteins, nucleic acids, and their multifaceted interactions within biological systems.
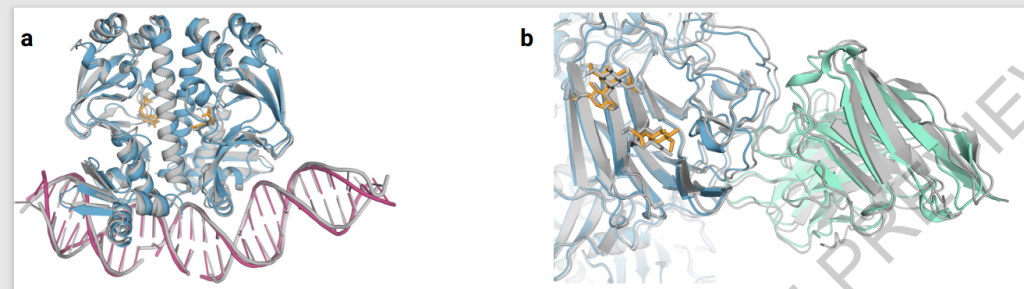
The primary obstacle in computational biology is the precise prediction of complex biomolecular structures, which is crucial for advancing fundamental biological understanding and enhancing therapeutic interventions. Predicting these complex interactions is integral to grasping cellular mechanisms and creating targeted treatments. Traditional computational models often fail to capture biomolecular systems’ true complexity and dynamics, significantly needing more sophisticated and accurate predictive tools.
Recent developments have improved predictive capabilities, yet they generally lack the accuracy required for complex molecular environments. While useful, the established models and tools often do not fully account for the intricate nature of molecular interactions, particularly in environments involving diverse types of molecules like proteins, nucleic acids, and small molecules.
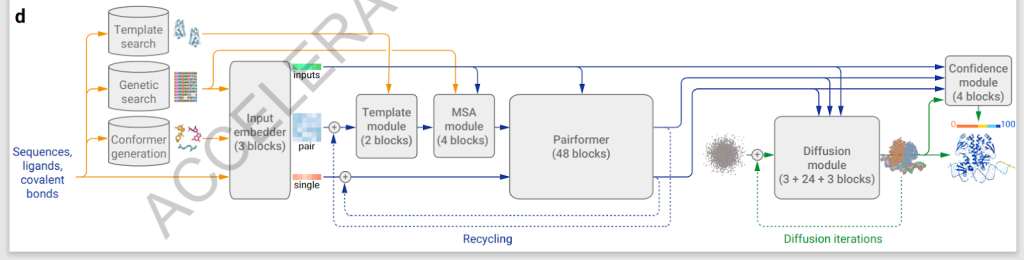
AlphaFold 3is a state-of-the-art tool from the Google DeepMind and Isomorphic Labs teams in computational biology for predicting the structure and interactions of all life’s molecules. This model employs a revolutionary diffusion-based architecture to enhance predictions’ accuracy significantly beyond existing tools’ capabilities. AlphaFold 3’s methodological advancements allow for comprehensive and precise modeling of biomolecular interactions that were previously unattainable with older computational techniques.
The model utilizes a novel approach by integrating a direct diffusion process that predicts raw atom coordinates, bypassing previous models’ limitations requiring detailed, often unavailable, experimental data. This approach has led to remarkable accuracy improvements in predicting the structure of protein complexes and interactions with small molecules and nucleic acids. For example, AlphaFold 3 achieves an interface accuracy of over 90% across various molecular interactions, substantially improving over traditional docking tools and other predictive models.
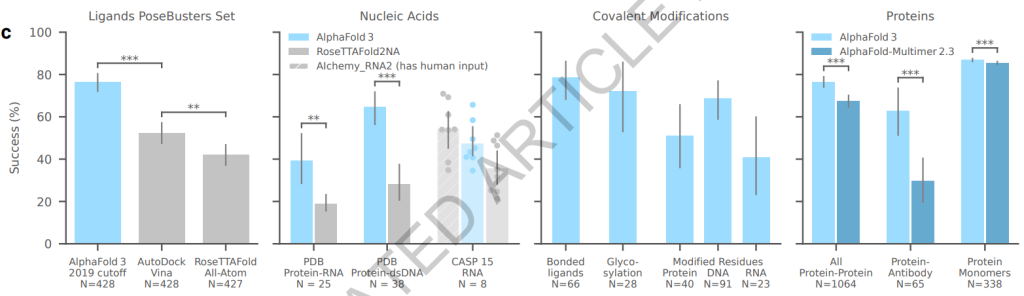
AlphaFold 3 demonstrates impressive performance metrics across various molecular interactions. It shows a Root Mean Square Deviation (RMSD) of less than 2 Å for most protein-ligand interactions tested, a clear enhancement compared to earlier models. The model performs exceptionally well in tests involving protein-nucleic acid interactions, with accuracy rates exceeding those of specialized nucleic acid predictors.
In conclusion, AlphaFold 3 is a monumental advance in biomolecular structure prediction, setting a new benchmark for accuracy and reliability in the field. Its ability to accurately model various biomolecular interactions opens new avenues for scientific research and drug development. By leveraging deep learning techniques to overcome traditional computational limits, AlphaFold 3 enhances the understanding of biological structures and accelerates the pace of biomedical discoveries, potentially leading to disease treatment and prevention breakthroughs. This tool’s success highlights the transformative impact of integrating advanced computational methods with biological research to tackle some of the most challenging problems in science today.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter.











